With the vision of P4 medicine (Predictive, Personalized, Preventive, Participatory), we innovate in the following areas:
- Developing analytical technologies for studying extracellular vesicles (EVs);
- Developing lab-on-chips for the discovery and validation of molecular biomarkers for health care and disease management in biofluids, cells, EVs, and tissue samples;
- The translation of these molecular biomarkers to clinical practices, for both laboratory tests and point-of-care applications;
- Developing biosensors for food safety applications.
Our research topics include:
Metal nanoparticle-enhanced 3D platforms for highly sensitive detection of EVs and proteins
Detection of diseases at early stages is critical to patient outcomes. Early disease diagnostics using circulating biomarkers requires highly sensitive biosensing technologies that can detect minimal amounts of molecules from complex biofluids such as blood plasma. We have developed metal nanomaterial (e.g., gold nanoparticles) enhanced 3D platforms with improved fluorescent signals for highly sensitive bioassays. The technology has been used to quantify multiple cancer-related proteins and exosomes in blood plasma samples, achieving up to four orders of magnitude improvement in sensitivity compared to conventional ELISA. The method is also multiplexed and easy to use, suitable for a wide range of bioassay applications.

Hydrogel-based biochemical assays
Hydrogels are hydrophilic polymers that can hold a large amount of water while maintaining their 3D structures. Their biocompatibility, high water content, and availability at a reasonable cost makes them widely used in tissue engineering, drug delivery, and food industry. We take advantage of the physical and chemical properties of some natural hydrogels to develop high-throughput biochemical assays for cancer research. These technologies will enable the establishment of personalized cancer diagnostics and will accelerate individualized treatment, offering a helping hand to cancer patients.
We are currently working on a novel hydrogel-based bioassay platform for sensitive cell analysis.
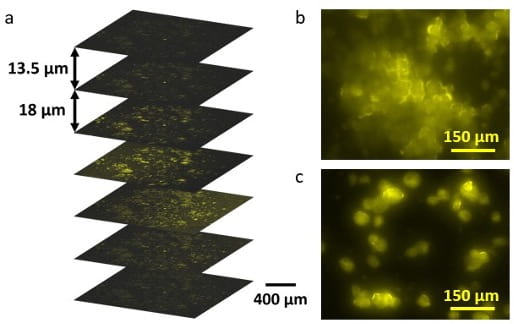
Scalable and transferrable microarrays for multiplexed bioassays
To conduct various physiological and pathological functions, in the human body, billions of biomolecules and trillions of cells work together and form complex networks. Therefore, multiplexed techniques are required to decipher the complexity of life. Microarray technology has been an essential tool for multiplexed bioassays and has been developed for decades. Although powerful, the process requires specialized skills and high-end instruments, limiting their broader applications. We have developed new formats of microarrays to conduct multiplexed bioassays without the need for complex instruments, making it widely available for researchers with basic lab setup and facilitating technology dissemination.
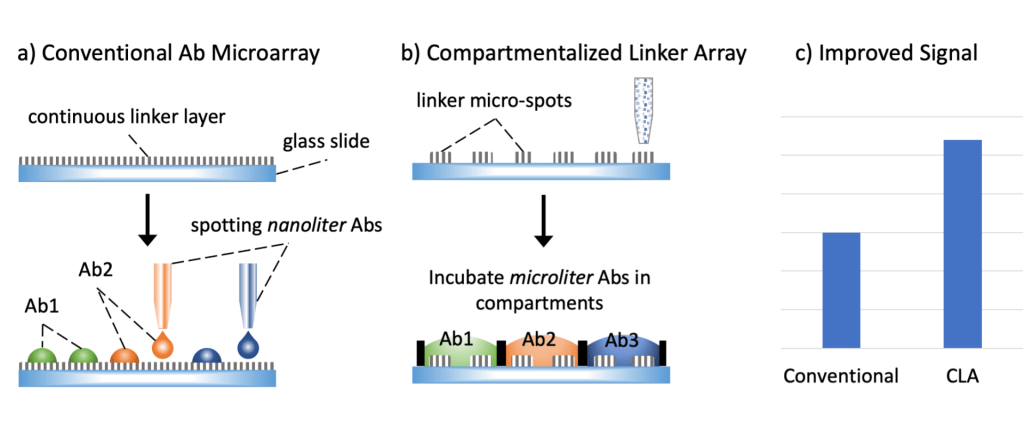
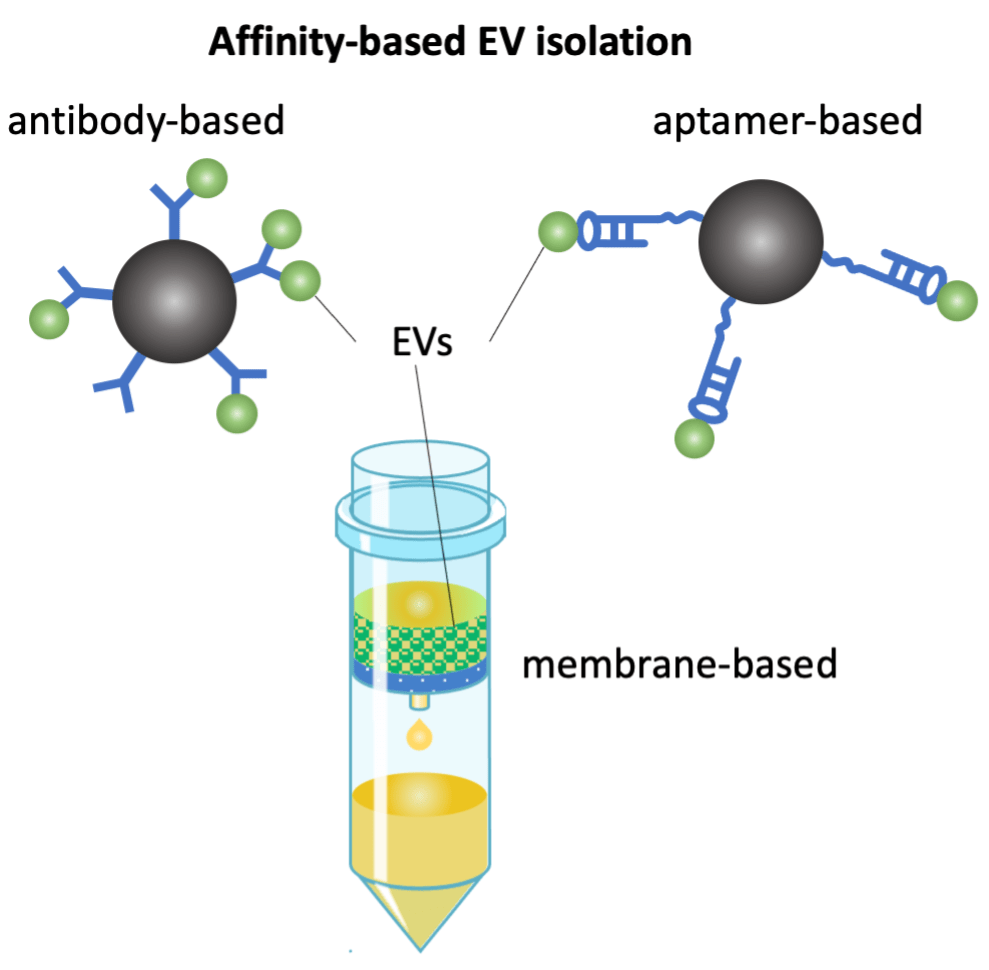
Affinity-based isolation of extracellular vesicles (EVs) from complex biofluids
Extracellular vesicles (EVs) are nanoscale particles secreted from cells. EVs carry biomolecular cargo very similar to their cells of origin, thus becoming a promising type of biomarker for disease management. Analysis of EV counting and their molecular composition from complex biofluids typically requires an EV isolation step. Current EV isolation methods face challenges in purity and yield, and are sometimes unsuitable with downstream molecular analysis. We are developing a novel affinity-based EV isolation, aiming to collect intact EVs with high purity and high yield, compatible with downstream analysis.
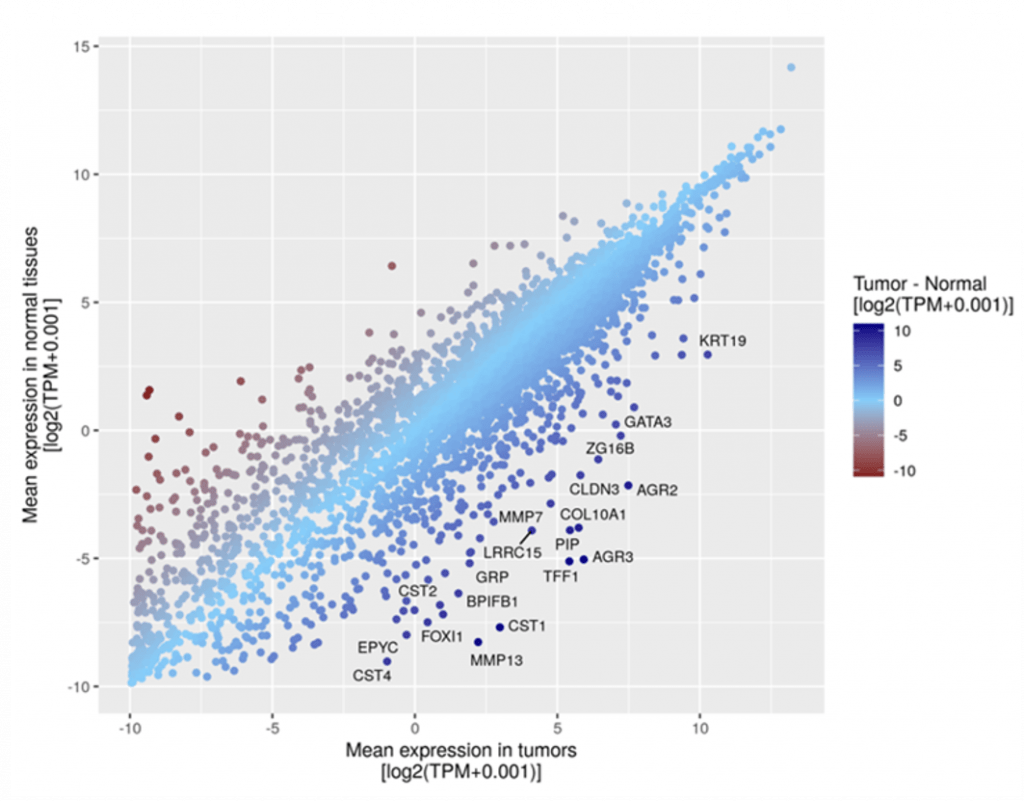
Computational tools for in-silico EV and protein cancer biomarker discovery
Biomarkers can be used in multiple stages of cancer management, such as diagnostics, prognosis, and treatment monitoring. Howver, discovering cancer biomarkers from vast amounts of biomolecules is a lengthy and costly journey. To accelerate the biomarker discovery process and reduce the workload required in labs, we have developed computational tools for in-silico selection of potential cancer biomarkers from publicly accessible OMICS databases with collective efforts from all over the world. Both genomics and proteomics data are investigated and compared for the biomarker selection. The shortlisted markers can potentially be further validated experimentally using clinical samples in labs.